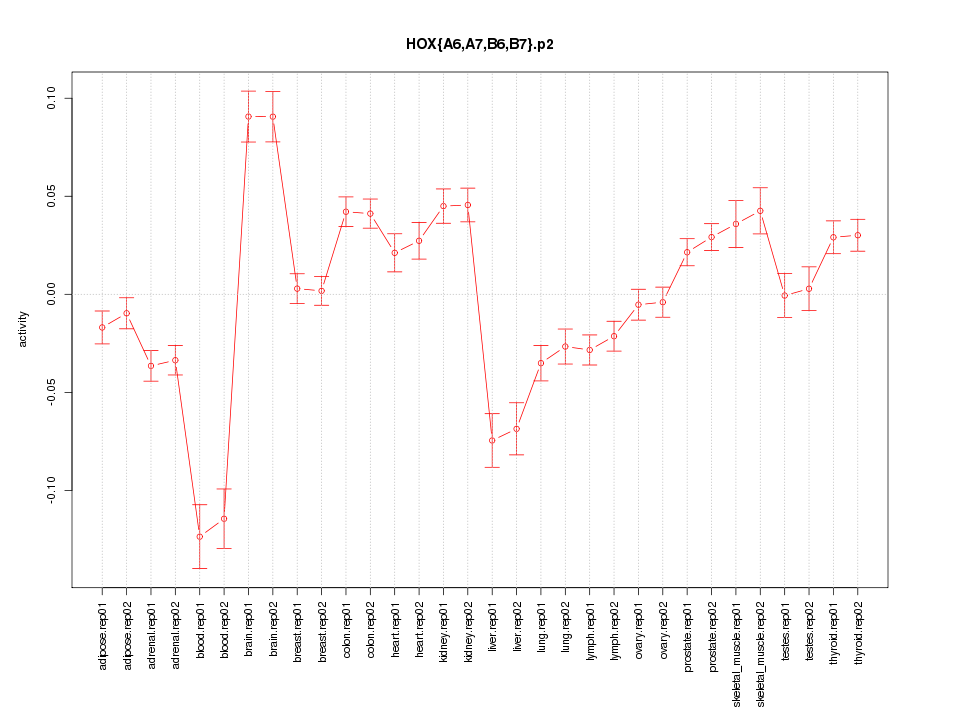
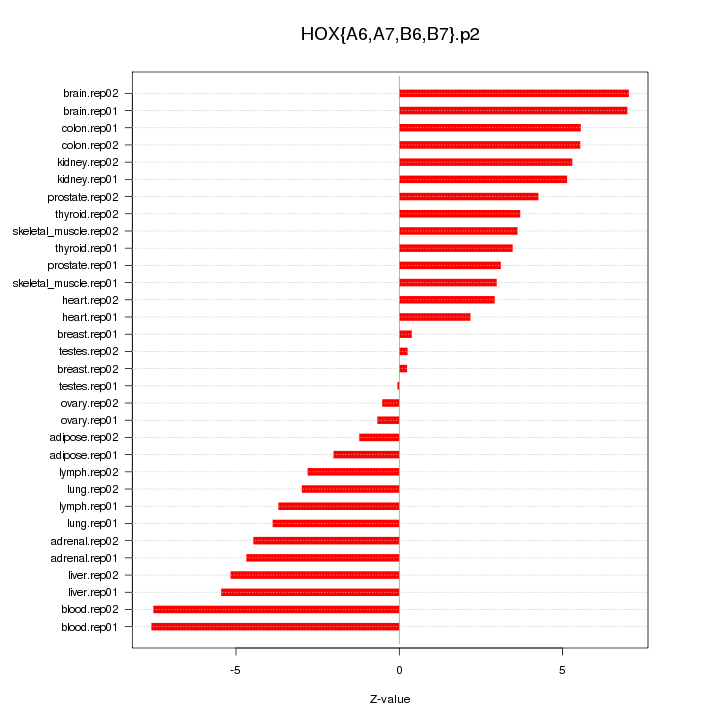
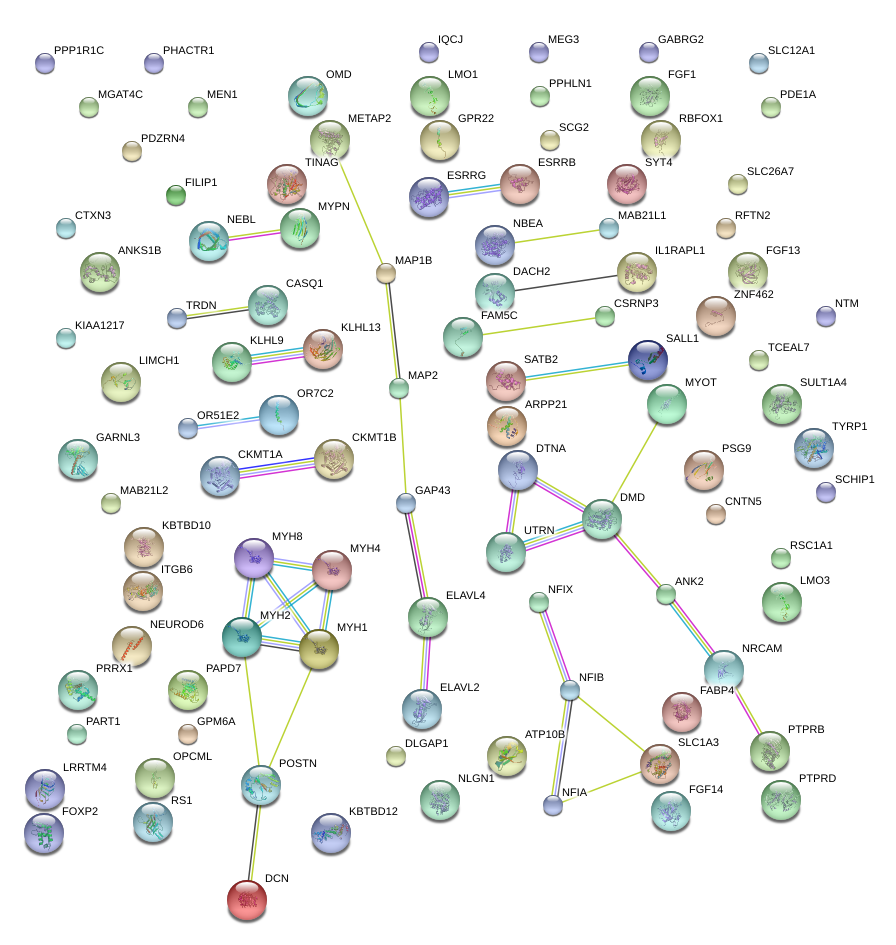
|
chr16_+_7382750
|
33.879
|
NM_145891
NM_145892
NM_145893
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr12_-_16760935
|
27.938
|
NM_001001395
NM_001243609
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr5_+_71502700
|
24.951
|
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr9_-_14086901
|
22.637
|
|
NFIB
|
nuclear factor I/B
|
|
chr12_-_87232635
|
20.389
|
NM_013244
|
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative)
|
|
chr3_+_159557745
|
20.177
|
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr1_-_217113007
|
19.395
|
NM_001243512
NM_001243513
NM_001243510
NM_001243511
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr1_-_217250230
|
18.910
|
NM_001243507
NM_001243509
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr3_+_127641901
|
18.183
|
NM_207335
|
KBTBD12
|
kelch repeat and BTB (POZ) domain containing 12
|
|
chr9_-_95186559
|
18.053
|
NM_005014
|
OMD
|
osteomodulin
|
|
chr2_+_170366211
|
17.810
|
NM_006063
|
KBTBD10
|
kelch repeat and BTB (POZ) domain containing 10
|
|
chr2_-_183387063
|
17.749
|
NM_005019
NM_001003683
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent
|
|
chr18_-_40857245
|
16.919
|
NM_020783
|
SYT4
|
synaptotagmin IV
|
|
chr3_+_35721115
|
16.334
|
NM_001025068
NM_001025069
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr3_+_159557649
|
16.103
|
NM_001197109
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr12_+_41831484
|
16.012
|
NM_013377
|
PDZRN4
|
PDZ domain containing ring finger 4
|
|
chr7_-_107880492
|
15.964
|
NM_001037132
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chr6_-_123957941
|
15.941
|
NM_001251987
NM_006073
|
TRDN
|
triadin
|
|
chrX_+_102585113
|
15.915
|
NM_152278
|
TCEAL7
|
transcription elongation factor A (SII)-like 7
|
|
chr3_+_158787040
|
15.629
|
NM_001197113
NM_001197114
NM_001042705
NM_001042706
NM_001197100
|
IQCJ-SCHIP1
IQCJ
|
IQCJ-SCHIP1 readthrough
IQ motif containing J
|
|
chr10_-_21186530
|
15.625
|
NM_006393
|
NEBL
|
nebulette
|
|
chr13_-_36050758
|
15.473
|
NM_005584
|
MAB21L1
|
mab-21-like 1 (C. elegans)
|
|
chr10_+_69869249
|
15.393
|
NM_032578
|
MYPN
|
myopalladin
|
|
chrX_-_137821514
|
15.355
|
NM_033642
|
FGF13
|
fibroblast growth factor 13
|
|
chr7_+_107110501
|
13.739
|
NM_005295
|
GPR22
|
G protein-coupled receptor 22
|
|
chr13_-_103054027
|
13.728
|
NM_175929
|
FGF14
|
fibroblast growth factor 14
|
|
chr4_-_176733413
|
13.618
|
|
GPM6A
|
glycoprotein M6A
|
|
chr5_+_59784004
|
13.581
|
|
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding)
|
|
chr1_+_170632274
|
13.490
|
|
PRRX1
|
paired related homeobox 1
|
|
chr9_-_8733893
|
13.287
|
NM_001040712
NM_001171025
NM_130391
NM_130392
NM_130393
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D
|
|
chr18_+_32290195
|
13.213
|
NM_001198941
NM_032978
|
DTNA
|
dystrobrevin, alpha
|
|
chr2_-_224467049
|
13.059
|
|
SCG2
|
secretogranin II
|
|
chr5_+_161494647
|
12.820
|
NM_000816
NM_198903
NM_198904
|
GABRG2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2
|
|
chr6_+_12716887
|
12.333
|
|
PHACTR1
|
phosphatase and actin regulator 1
|
|
chr4_+_41614911
|
12.279
|
NM_001112719
NM_001112720
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr2_+_210444364
|
12.068
|
NM_002374
NM_031845
NM_031847
|
MAP2
|
microtubule-associated protein 2
|
|
chr4_+_41614725
|
12.012
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr14_+_101295409
|
11.997
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr11_+_99426872
|
11.796
|
NM_175566
|
CNTN5
|
contactin 5
|
|
chr6_-_76203256
|
11.776
|
NM_015687
|
FILIP1
|
filamin A interacting protein 1
|
|
chr4_-_176733378
|
11.648
|
|
GPM6A
|
glycoprotein M6A
|
|
chr18_+_32173281
|
11.590
|
NM_001128175
NM_001198938
NM_001198939
NM_001198940
|
DTNA
|
dystrobrevin, alpha
|
|
chr9_+_109686436
|
11.480
|
|
ZNF462
|
zinc finger protein 462
|
|
chr2_-_198540583
|
11.327
|
NM_144629
|
RFTN2
|
raftlin family member 2
|
|
chr18_-_3845295
|
11.206
|
NM_001003809
NM_001242765
NM_001242766
|
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1
|
|
chr10_+_24755459
|
11.121
|
|
KIAA1217
|
KIAA1217
|
|
chr4_+_113970784
|
10.993
|
NM_001148
NM_020977
|
ANK2
|
ankyrin 2, neuronal
|
|
chr11_-_4718948
|
10.858
|
NM_030774
|
OR51E2
|
olfactory receptor, family 51, subfamily E, member 2
|
|
chr4_+_41614933
|
10.748
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr17_-_10452939
|
10.677
|
NM_001100112
NM_017534
|
MYH2
|
myosin, heavy chain 2, skeletal muscle, adult
|
|
chr6_+_54173202
|
10.666
|
NM_014464
|
TINAG
|
tubulointerstitial nephritis antigen
|
|
chr12_-_99548867
|
10.652
|
NM_001204067
|
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B
|
|
chr9_+_12693385
|
10.483
|
NM_000550
|
TYRP1
|
tyrosinase-related protein 1
|
|
chr9_+_130026755
|
10.400
|
NM_032293
|
GARNL3
|
GTPase activating Rap/RanGAP domain-like 3
|
|
chr2_-_200325200
|
10.283
|
|
SATB2
|
SATB homeobox 2
|
|
chr17_-_10421858
|
10.115
|
NM_005963
|
MYH1
|
myosin, heavy chain 1, skeletal muscle, adult
|
|
chr12_-_16758312
|
10.094
|
NM_001243611
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr11_+_131240370
|
10.076
|
NM_001048209
|
NTM
|
neurotrimin
|
|
chr4_+_41700736
|
10.024
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr4_-_176733901
|
9.907
|
|
GPM6A
|
glycoprotein M6A
|
|
chr12_-_91576805
|
9.855
|
NM_001920
|
DCN
|
decorin
|
|
chrX_-_32430370
|
9.744
|
NM_004011
NM_004012
|
DMD
|
dystrophin
|
|
chrX_+_85517970
|
9.690
|
NM_001139515
|
DACH2
|
dachshund homolog 2 (Drosophila)
|
|
chr1_+_160160284
|
9.682
|
NM_001231
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle)
|
|
chr12_-_16758839
|
9.653
|
NM_001243612
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chrX_-_18690187
|
9.652
|
NM_000330
|
RS1
|
retinoschisin 1
|
|
chr2_-_77749335
|
9.483
|
NM_001134745
NM_024993
|
LRRTM4
|
leucine rich repeat transmembrane neuronal 4
|
|
chr7_-_31380507
|
9.442
|
NM_022728
|
NEUROD6
|
neurogenic differentiation 6
|
|
chr2_-_224467095
|
9.431
|
|
SCG2
|
secretogranin II
|
|
chr5_+_126984712
|
9.398
|
NM_001048252
|
CTXN3
|
cortexin 3
|
|
chr5_+_137203559
|
9.226
|
|
MYOT
|
myotilin
|
|
chr14_+_76837689
|
9.141
|
NM_004452
|
ESRRB
|
estrogen-related receptor beta
|
|
chr8_+_92261515
|
9.054
|
NM_052832
NM_134266
|
SLC26A7
|
solute carrier family 26, member 7
|
|
chr3_+_173116243
|
9.032
|
NM_014932
|
NLGN1
|
neuroligin 1
|
|
chr5_-_141993691
|
8.959
|
NM_033137
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr2_+_166428845
|
8.927
|
NM_024969
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3
|
|
chr5_+_36606667
|
8.910
|
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr15_+_48498497
|
8.871
|
NM_000338
NM_001184832
|
SLC12A1
|
solute carrier family 12 (sodium/potassium/chloride transporters), member 1
|
|
chr1_+_50574601
|
8.854
|
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr12_-_71031174
|
8.830
|
NM_001109754
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B
|
|
chr8_-_82395401
|
8.735
|
NM_001442
|
FABP4
|
fatty acid binding protein 4, adipocyte
|
|
chr2_-_161056762
|
8.720
|
|
ITGB6
|
integrin, beta 6
|
|
chr17_+_22022436
|
8.702
|
NM_001190452
|
MTRNR2L1
|
MT-RNR2-like 1
|
|
chrX_+_28605558
|
8.659
|
NM_014271
|
IL1RAPL1
|
interleukin 1 receptor accessory protein-like 1
|
|
chr2_+_182850550
|
8.618
|
NM_001080545
|
PPP1R1C
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C
|
|
chr2_-_224467142
|
8.554
|
NM_003469
|
SCG2
|
secretogranin II
|
|
chr12_-_16759430
|
8.549
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr13_-_38172862
|
8.524
|
NM_001135934
NM_001135935
NM_001135936
NM_006475
|
POSTN
|
periostin, osteoblast specific factor
|
|
chr5_+_36606456
|
8.493
|
NM_001166696
NM_004172
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chrX_-_117119282
|
8.466
|
NM_001168299
|
KLHL13
|
kelch-like 13 (Drosophila)
|
|
chr13_+_36050885
|
8.452
|
NM_001204197
|
NBEA
|
neurobeachin
|
|
chr3_+_115342302
|
8.443
|
|
GAP43
|
growth associated protein 43
|
|
chr7_+_114055234
|
8.428
|
|
FOXP2
|
forkhead box P2
|
|
chr5_+_137203544
|
8.416
|
NM_001135940
NM_006790
|
MYOT
|
myotilin
|
|
chr18_+_32073245
|
8.395
|
NM_001198945
NM_001392
NM_032975
NM_032979
|
DTNA
|
dystrobrevin, alpha
|
|
chr15_+_43885251
|
8.373
|
NM_020990
|
CKMT1B
CKMT1A
|
creatine kinase, mitochondrial 1B
creatine kinase, mitochondrial 1A
|
|
chr5_-_160279045
|
8.322
|
NM_025153
|
ATP10B
|
ATPase, class V, type 10B
|
|
chr12_-_16759733
|
8.272
|
NM_001243610
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr16_-_51185150
|
8.220
|
NM_002968
|
SALL1
|
sal-like 1 (Drosophila)
|
|
chr1_-_190446701
|
8.212
|
NM_199051
|
FAM5C
|
family with sequence similarity 5, member C
|
|
chr9_-_95166827
|
8.206
|
NM_014057
NM_033014
|
OGN
|
osteoglycin
|
|
chr1_-_216896640
|
8.176
|
NM_001243515
NM_001243518
NM_001243519
NM_001438
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr10_+_24755440
|
8.174
|
|
KIAA1217
|
KIAA1217
|
|
chr5_+_139505519
|
8.145
|
NM_001007189
|
IGIP
|
IgA-inducing protein homolog (Bos taurus)
|
|
chr21_-_39870303
|
8.138
|
NM_001136155
NM_182918
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr12_-_15865847
|
8.088
|
|
EPS8
|
epidermal growth factor receptor pathway substrate 8
|
|
chr3_+_63428752
|
8.076
|
NM_144642
|
SYNPR
|
synaptoporin
|
|
chr18_-_53089722
|
7.959
|
NM_001243233
|
TCF4
|
transcription factor 4
|
|
chr17_+_68071386
|
7.948
|
NM_018658
NM_170741
|
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16
|
|
chr12_-_91576579
|
7.914
|
|
DCN
|
decorin
|
|
chr3_+_135741576
|
7.885
|
NM_181897
|
PPP2R3A
|
protein phosphatase 2, regulatory subunit B'', alpha
|
|
chr10_+_45406629
|
7.862
|
NM_001123376
|
TMEM72
|
transmembrane protein 72
|
|
chrX_+_135278912
|
7.853
|
NM_001159701
NM_001159699
|
FHL1
|
four and a half LIM domains 1
|
|
chr3_+_181417387
|
7.848
|
|
SOX2-OT
|
SOX2 overlapping transcript (non-protein coding)
|
|
chr10_-_69455926
|
7.814
|
NM_013266
|
CTNNA3
|
catenin (cadherin-associated protein), alpha 3
|
|
chr12_-_16759420
|
7.798
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr18_-_64271215
|
7.794
|
NM_021153
|
CDH19
|
cadherin 19, type 2
|
|
chr12_-_16759531
|
7.786
|
NM_018640
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr22_-_37098620
|
7.776
|
NM_006078
|
CACNG2
|
calcium channel, voltage-dependent, gamma subunit 2
|
|
chr13_-_36429798
|
7.758
|
NM_001195415
NM_001195416
NM_001195430
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr3_-_69171536
|
7.738
|
NM_198271
|
LMOD3
|
leiomodin 3 (fetal)
|
|
chr4_-_186877869
|
7.736
|
NM_021069
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr9_+_103340335
|
7.703
|
NM_001018116
|
MURC
|
muscle-related coiled-coil protein
|
|
chr3_-_167191808
|
7.684
|
NM_006217
|
SERPINI2
|
serpin peptidase inhibitor, clade I (pancpin), member 2
|
|
chr15_+_71839540
|
7.659
|
|
THSD4
|
thrombospondin, type I, domain containing 4
|
|
chr14_+_62462540
|
7.585
|
NM_031914
|
SYT16
|
synaptotagmin XVI
|
|
chr4_+_113739203
|
7.529
|
NM_001127493
|
ANK2
|
ankyrin 2, neuronal
|
|
chr8_+_80523376
|
7.526
|
|
STMN2
|
stathmin-like 2
|
|
chr10_+_68685791
|
7.513
|
NM_178011
|
LRRTM3
|
leucine rich repeat transmembrane neuronal 3
|
|
chr2_-_161056573
|
7.494
|
NM_000888
|
ITGB6
|
integrin, beta 6
|
|
chr3_+_35722428
|
7.460
|
NM_198399
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr12_-_16757944
|
7.456
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr8_+_104831415
|
7.407
|
NM_014677
|
RIMS2
|
regulating synaptic membrane exocytosis 2
|
|
chr18_+_59157774
|
7.385
|
NM_031891
|
CDH20
|
cadherin 20, type 2
|
|
chr9_-_19786886
|
7.322
|
NM_001193288
NM_020344
|
SLC24A2
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 2
|
|
chr4_-_186733372
|
7.275
|
NM_003603
NM_001145670
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr5_-_96478502
|
7.218
|
NM_153234
|
LIX1
|
Lix1 homolog (chicken)
|
|
chr18_-_3874252
|
7.215
|
NM_001242764
|
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1
|
|
chr1_-_203055088
|
7.209
|
|
MYOG
|
myogenin (myogenic factor 4)
|
|
chr10_+_95517565
|
7.153
|
NM_005097
|
LGI1
|
leucine-rich, glioma inactivated 1
|
|
chr11_+_101983170
|
7.126
|
NM_001195045
|
YAP1
|
Yes-associated protein 1
|
|
chr16_+_6823809
|
7.124
|
NM_001142334
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr5_-_24645084
|
7.123
|
NM_006727
|
CDH10
|
cadherin 10, type 2 (T2-cadherin)
|
|
chrX_-_32173585
|
7.113
|
NM_004013
NM_004020
NM_004021
NM_004022
NM_004023
|
DMD
|
dystrophin
|
|
chr5_-_124080773
|
7.030
|
NM_020747
|
ZNF608
|
zinc finger protein 608
|
|
chr22_-_36236421
|
6.982
|
NM_001031695
NM_001082576
NM_001082577
NM_014309
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chrX_-_13835313
|
6.966
|
NM_001001995
NM_001001996
NM_005278
|
GPM6B
|
glycoprotein M6B
|
|
chr1_+_215178884
|
6.966
|
NM_001017424
|
KCNK2
|
potassium channel, subfamily K, member 2
|
|
chr4_+_88896801
|
6.956
|
NM_000582
NM_001040058
NM_001040060
NM_001251829
NM_001251830
|
SPP1
|
secreted phosphoprotein 1
|
|
chr5_+_166711842
|
6.954
|
NM_001122679
|
ODZ2
|
odz, odd Oz/ten-m homolog 2 (Drosophila)
|
|
chr2_-_233641225
|
6.924
|
NM_001172416
NM_001172417
NM_002242
|
KCNJ13
|
potassium inwardly-rectifying channel, subfamily J, member 13
|
|
chr12_-_117799410
|
6.922
|
NM_000620
NM_001204218
|
NOS1
|
nitric oxide synthase 1 (neuronal)
|
|
chr4_-_21699317
|
6.908
|
NM_001035003
|
KCNIP4
|
Kv channel interacting protein 4
|
|
chr10_-_75335415
|
6.873
|
NM_152586
|
USP54
|
ubiquitin specific peptidase 54
|
|
chr11_+_113775517
|
6.866
|
NM_006028
|
HTR3B
|
5-hydroxytryptamine (serotonin) receptor 3B
|
|
chr4_-_87770267
|
6.857
|
NM_197965
|
SLC10A6
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 6
|
|
chr6_+_121756744
|
6.851
|
NM_000165
|
GJA1
|
gap junction protein, alpha 1, 43kDa
|
|
chr3_+_25470067
|
6.848
|
|
RARB
|
retinoic acid receptor, beta
|
|
chr4_+_71494460
|
6.827
|
NM_031889
|
ENAM
|
enamelin
|
|
chr11_+_27015668
|
6.783
|
|
|
|
|
chr5_+_174151531
|
6.755
|
NM_002449
|
MSX2
|
msh homeobox 2
|
|
chr13_+_96085852
|
6.718
|
NM_001160100
NM_182848
|
CLDN10
|
claudin 10
|
|
chr1_-_178840079
|
6.679
|
NM_004673
|
ANGPTL1
|
angiopoietin-like 1
|
|
chr11_-_8285299
|
6.667
|
NM_002315
|
LMO1
|
LIM domain only 1 (rhombotin 1)
|
|
chr11_+_27015627
|
6.639
|
NM_203371
|
FIBIN
|
fin bud initiation factor homolog (zebrafish)
|
|
chr17_-_10372875
|
6.617
|
NM_017533
|
MYH4
|
myosin, heavy chain 4, skeletal muscle
|
|
chr2_-_105467912
|
6.610
|
|
LOC100506421
|
uncharacterized LOC100506421
|
|
chr4_-_186877413
|
6.596
|
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr10_+_95848811
|
6.565
|
NM_001165979
|
PLCE1
|
phospholipase C, epsilon 1
|
|
chr12_+_119616594
|
6.560
|
NM_014365
|
HSPB8
|
heat shock 22kDa protein 8
|
|
chr3_+_2280512
|
6.548
|
NM_001206955
|
CNTN4
|
contactin 4
|
|
chr4_+_88896838
|
6.522
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr11_-_111782447
|
6.518
|
NM_001885
|
CRYAB
|
crystallin, alpha B
|
|
chr11_-_115088628
|
6.502
|
|
CADM1
|
cell adhesion molecule 1
|
|
chr10_-_62149487
|
6.492
|
NM_020987
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr1_+_81771844
|
6.485
|
|
LPHN2
|
latrophilin 2
|
|
chr1_+_65730423
|
6.469
|
NM_014787
|
DNAJC6
|
DnaJ (Hsp40) homolog, subfamily C, member 6
|
|
chr7_+_114055048
|
6.325
|
NM_001172766
NM_014491
NM_148898
NM_148900
|
FOXP2
|
forkhead box P2
|
|
chr18_+_616671
|
6.321
|
NM_199167
|
CLUL1
|
clusterin-like 1 (retinal)
|
|
chr4_+_88896868
|
6.312
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr8_+_77593522
|
6.254
|
|
ZFHX4
|
zinc finger homeobox 4
|
|
chr8_+_80523351
|
6.251
|
|
STMN2
|
stathmin-like 2
|
|
chr18_-_31628557
|
6.236
|
NM_001198549
|
NOL4
|
nucleolar protein 4
|
|
chr13_-_70682458
|
6.235
|
NM_020866
|
KLHL1
|
kelch-like 1 (Drosophila)
|
|
chr6_-_46293628
|
6.235
|
NM_005822
|
RCAN2
|
regulator of calcineurin 2
|
|
chrX_-_13835187
|
6.163
|
|
GPM6B
|
glycoprotein M6B
|
|
chr11_-_26743545
|
6.162
|
NM_178498
|
SLC5A12
|
solute carrier family 5 (sodium/glucose cotransporter), member 12
|
|
chr3_+_42190825
|
6.156
|
|
TRAK1
|
trafficking protein, kinesin binding 1
|
|
chr5_-_96478438
|
6.144
|
|
LIX1
|
Lix1 homolog (chicken)
|
|
chr2_+_210444154
|
6.144
|
|
MAP2
|
microtubule-associated protein 2
|
|
chr6_-_24383519
|
6.099
|
NM_001195610
|
DCDC2
|
doublecortin domain containing 2
|
|
chr22_-_36236264
|
6.083
|
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr6_-_116381765
|
6.080
|
NM_002031
|
FRK
|
fyn-related kinase
|
|
chr8_+_77593506
|
6.067
|
NM_024721
|
ZFHX4
|
zinc finger homeobox 4
|
|
chr14_+_32798478
|
6.062
|
NM_004274
|
AKAP6
|
A kinase (PRKA) anchor protein 6
|
|
chrX_-_31526414
|
6.020
|
NM_004014
|
DMD
|
dystrophin
|
|
chr17_-_29624249
|
5.996
|
NM_002544
|
OMG
|
oligodendrocyte myelin glycoprotein
|
|
chr1_+_177140062
|
5.989
|
|
FAM5B
|
family with sequence similarity 5, member B
|
|
chr4_+_88896899
|
5.989
|
|
SPP1
|
secreted phosphoprotein 1
|
|
chr8_+_80523048
|
5.988
|
NM_001199214
NM_007029
|
STMN2
|
stathmin-like 2
|